


The request for calculation is send and then on the Cytoscape side forgotten about. The Cytoscape NetworkAnalyzer results are rounded, whereas the NeoNetworkAnalyzer returns the unrounded double values.Īsynchronous Execution is intended for large network in the Neo4j instance. You can calculate the same results using Cytoscape NetworkAnalyzer at Tools → NetworkAnalyzer → Network Analysis → Analyze Network.CyNeo4j adds those as properties with the prefix neo_ to the Cytoscape network. As soon as the calculation is finished the Neo4j instance will return the results.Click OK and wait for the calculation to finish. Leave the Asynchronous Execution unticked. Tick the Save in Graph option to store the results also in the Neo4j instance. By default the implementation calculates all node statistics.Launch the NeoNetworkAnalyzer via Apps → CyNeo4j → Statistics → NeoNetworkAnalyzer.Load it into a running Neo4j instance with Sync Up.Open the example network ( download) in Cytoscape.(see Installation Instructions for more details) Start a Neo4j instance (if not running already) with the NeoNetworkAnalyzer plugin installed.The NeoNetworkAnalyzer is a Neo4j based reimplementation of the NetworkAnalyzer included in Cytoscape. Be careful if you have a large network stored there! The query downloads the whole graph from the Neo4j instance.The predefined query in the dialog is match (n)->(m) return n,r,m.The Cypher query interface is available (after connecting) at Apps → cyNeo4j → Cypher Query.Create a new empty network through File → New → Network → Empty Network.If you are not connected to a Neo4j instance yet do that!.Statistics like the result from a count operation are not supported in a meaningful way yet. As of version 1.0 of cyNeo4j the app can deal with Cypher queries that return node and edge objects.
We will focus here on what cyNeo4j can do with Cypher queries currently. Cypherįor an in depth tutorial on Cypher have a look at the official one of Neo4j here. Large networks like STRING can be too much for Cytoscape at once, so let us have a look at Cypher a query language develop by the team behind Neo4j. The Sync Down functionality obviously does the opposite but keep in mind that Neo4j can deal with networks far larger than Cytoscape can handle (on a typical desktop machine). This upload step can take quite a bit for bigger networks. The Sync Up functionality is intended to get a network into an empty Neo4j instance to use algorithms available on the instance. The old network there will be deleted! Note that the neoid column will change, as the new network has different ids on the Neo4j instance!
Use the Apps → cyNeo4j → Sync Up to upload your modified network to the Neo4j instance.
#Neo4j cytoscape movie
#Neo4j cytoscape download
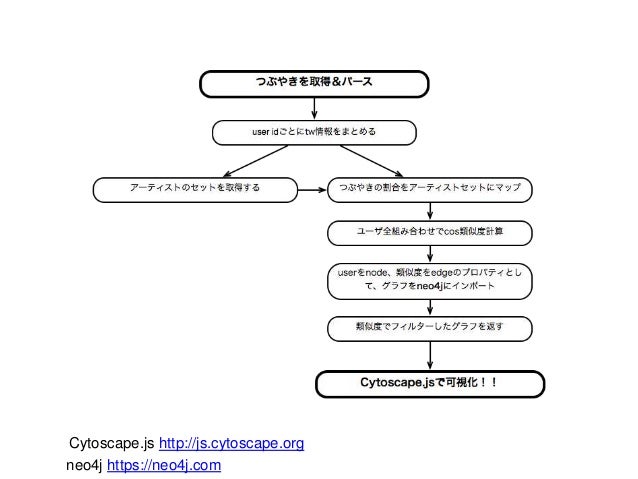
Download the movie graph from the Neo4j instance via Apps → cyNeo4j → Sync Down.Then load data and make network structure as json.ĭf=pd.read_table( "bioactivity-14_6-07-32.txt", header=0 ) If you interested in it, please read following -).Īt first, I get sample data from Chembl.(Cyclin-dependent kinase 2) It is an open-source JavaScript graph theory library for analysis and visualisation. Lots of relationships are presented as graph.įor example, if I set molecule as node, and similarity as edge, I could make molecular networks.


 0 kommentar(er)
0 kommentar(er)
